© Universität Bielefeld
uni.news
Published on
21. Dezember 2012
Category
General
A giant puzzle with billions of pieces
Bielefeld’s Center for Biotechnology and the Joint Genome Institute, USA, decipher genetic information of microbes in biogas plants
Day after day, legions of microorganisms work to produce energy from waste in biogas plants. Researchers from Bielefeld University’s Center for Biotechnology (CeBiTec) are taking a close look to find out which microbes do the best job. They are analysing the entire genetic information of the microbial communities in selected biogas plants up and down Germany. From the beginning of 2013, the Californian Joint Genome Institute will undertake the sequencing required. The biocomputational analysis will be performed at CeBiTec. Not an easy task, since the data will be supplied in billions of fragments stemming in turn from hundreds of organisms. Piecing together this huge jigsaw puzzle will be painstaking work.
 In Germany, there are more than 7,000 biogas plants which can supply over six million households with power. The plants are filled mostly with plant biomass like maize silage but also with agricultural waste materials like liquid manure and chicken manure. One of the key research questions is how the production of biogas can be optimised. For this reason, Bielefeld scientists Dr Alexander Sczyrba, Dr Andreas Schlüter, Dr Alexander Goesmann, Professor Dr Jens Stoye und Professor Dr Alfred Pühler want to know what microbes are responsible for the decomposition of biomass - and which of them do it best. “We are interested in discovering the microbiology that is really behind the processes going on in a biogas plant; what microorganisms play which role at which stage,” explains Andreas Schlüter, whose research at CeBiTec is in the field of biogas production.
In Germany, there are more than 7,000 biogas plants which can supply over six million households with power. The plants are filled mostly with plant biomass like maize silage but also with agricultural waste materials like liquid manure and chicken manure. One of the key research questions is how the production of biogas can be optimised. For this reason, Bielefeld scientists Dr Alexander Sczyrba, Dr Andreas Schlüter, Dr Alexander Goesmann, Professor Dr Jens Stoye und Professor Dr Alfred Pühler want to know what microbes are responsible for the decomposition of biomass - and which of them do it best. “We are interested in discovering the microbiology that is really behind the processes going on in a biogas plant; what microorganisms play which role at which stage,” explains Andreas Schlüter, whose research at CeBiTec is in the field of biogas production.
First genome deciphered
The researchers’ work has already borne its first fruit. “At CeBiTec, we have managed to decipher the complete genome sequence of Methanoculleus bourgensis, a methane producer,” reports Professor Pühler. By doing so, Bielefeld has sequenced the first genome for a methane-producing archaeon from a biogas plant – a single-celled primordial bacterium which plays an important role in certain biogas plants. Now, the researchers want to go even further.

Putting the puzzle together
The project is part of the Community Sequencing Program, a public sequencing programme financed at the Joint Genome Institute by the US Department of Energy. While previous biogas studies have concentrated primarily on certain marker genes, now the entire genetic information of the microorganisms is to be studied. The American institute will produce more than one terabyte of sequence data for this, which is equivalent in volume to approximately 300 human genomes. This data will be supplied in a countless number of fragments, however, since even the most modern technology is not capable of reading all at once the millions of bases of which a microbial DNA molecule consists. Instead, the sequencing technologies supply vast quantities of overlapping sections of about 150 bases. The DNA sequences will then be returned to Bielefeld in billions of fragments, which is where Alexander Sczyrba’s Computa-tional Metagenomics team comes into play. They develop bioinformatic procedures for the reconstruction of genome sequences. Their task is to compare the data, recognise the overlaps and use them to reassemble the base sequence. “We are trying to complete a puzzle made up of billions of pieces, which also includes hundreds of different puzzles all mixed up,” explains Sczyrba.
Single-cell genomics promises new insights
Quite incidentally, the Bielefeld researchers will be breaking new ground in genomics. An estimated 99 per cent of all microorganisms cannot be cultivated in the laboratory. A brand new technology, single-cell genomics, is to provide insights here by determining the genome sequence from single microbial cells. Knowledge of the identity and functions of hitherto completely unknown microorganisms is expected to be gained. During the joint project, the Joint Genome Institute will sequence approximately 100 single-cell genomes.
The researchers have scheduled roughly two years for their project, in which also Bielefeld doctoral students of the Graduate Cluster in Industrial Biotechnology (CLIB) are involved. At the end, they hope to have discovered the optimal microbial community for biogas plants - and thus be in a position to make this process of generating energy even more efficient.
Background
Biogas plants produce methane through the fermentation of plant biomass, which can be used to generate power and heat. The decomposition of plant biomass and the production of biogas in agricultural biogas plants are brought about by microbes. This process, which is similar to what goes on in the digestive tract of cattle, has a neutral carbon dioxide balance and does not therefore contribute to global warming. Unlike other renewable energies, for example weather-dependent power sources like wind and solar, methane can be produced constantly and stored. This allows it to be converted into power or heat as required.
For further information in the Internet, go to:
www.cebitec.uni-bielefeld.de/grim/index.php/research/metagenomes
Contact:
Dr Alexander Sczyrba, Bielefeld University
CeBiTec / Faculty of Technology - Computational Metagenomics
Telephone: +49 521 106-2910
Email: asczyrba@cebitec.uni-bielefeld.de
Dr Andreas Schlüter, Bielefeld University
CeBiTec - Genome Research of Industrial Microorganisms
Telephone: +49 521 106-8757
Email: aschluet@cebitec.uni-bielefeld.de
Day after day, legions of microorganisms work to produce energy from waste in biogas plants. Researchers from Bielefeld University’s Center for Biotechnology (CeBiTec) are taking a close look to find out which microbes do the best job. They are analysing the entire genetic information of the microbial communities in selected biogas plants up and down Germany. From the beginning of 2013, the Californian Joint Genome Institute will undertake the sequencing required. The biocomputational analysis will be performed at CeBiTec. Not an easy task, since the data will be supplied in billions of fragments stemming in turn from hundreds of organisms. Piecing together this huge jigsaw puzzle will be painstaking work.
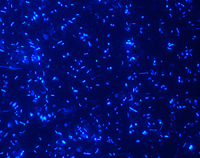
Stained, they fluoresce under the microscope: a wide variety of microbes can be seen in a sample taken from a biogas plant. Researchers at Bielefeld University want to find out which ones do their job best. Image: Karsten Niehaus
First genome deciphered
The researchers’ work has already borne its first fruit. “At CeBiTec, we have managed to decipher the complete genome sequence of Methanoculleus bourgensis, a methane producer,” reports Professor Pühler. By doing so, Bielefeld has sequenced the first genome for a methane-producing archaeon from a biogas plant – a single-celled primordial bacterium which plays an important role in certain biogas plants. Now, the researchers want to go even further.
Bielefeld researchers visit the site of a Bielefeld biogas plant (from left): Dr Andreas Schlüter, Prof. Dr Jens Stoye, Dr Alexander Goesmann, Dr Alexander Sczyrba und Prof. Dr Alfred Pühler. Photo: Björn Fischer
Putting the puzzle together
The project is part of the Community Sequencing Program, a public sequencing programme financed at the Joint Genome Institute by the US Department of Energy. While previous biogas studies have concentrated primarily on certain marker genes, now the entire genetic information of the microorganisms is to be studied. The American institute will produce more than one terabyte of sequence data for this, which is equivalent in volume to approximately 300 human genomes. This data will be supplied in a countless number of fragments, however, since even the most modern technology is not capable of reading all at once the millions of bases of which a microbial DNA molecule consists. Instead, the sequencing technologies supply vast quantities of overlapping sections of about 150 bases. The DNA sequences will then be returned to Bielefeld in billions of fragments, which is where Alexander Sczyrba’s Computa-tional Metagenomics team comes into play. They develop bioinformatic procedures for the reconstruction of genome sequences. Their task is to compare the data, recognise the overlaps and use them to reassemble the base sequence. “We are trying to complete a puzzle made up of billions of pieces, which also includes hundreds of different puzzles all mixed up,” explains Sczyrba.
Single-cell genomics promises new insights
Quite incidentally, the Bielefeld researchers will be breaking new ground in genomics. An estimated 99 per cent of all microorganisms cannot be cultivated in the laboratory. A brand new technology, single-cell genomics, is to provide insights here by determining the genome sequence from single microbial cells. Knowledge of the identity and functions of hitherto completely unknown microorganisms is expected to be gained. During the joint project, the Joint Genome Institute will sequence approximately 100 single-cell genomes.
The researchers have scheduled roughly two years for their project, in which also Bielefeld doctoral students of the Graduate Cluster in Industrial Biotechnology (CLIB) are involved. At the end, they hope to have discovered the optimal microbial community for biogas plants - and thus be in a position to make this process of generating energy even more efficient.
Background
Biogas plants produce methane through the fermentation of plant biomass, which can be used to generate power and heat. The decomposition of plant biomass and the production of biogas in agricultural biogas plants are brought about by microbes. This process, which is similar to what goes on in the digestive tract of cattle, has a neutral carbon dioxide balance and does not therefore contribute to global warming. Unlike other renewable energies, for example weather-dependent power sources like wind and solar, methane can be produced constantly and stored. This allows it to be converted into power or heat as required.
For further information in the Internet, go to:
www.cebitec.uni-bielefeld.de/grim/index.php/research/metagenomes
Contact:
Dr Alexander Sczyrba, Bielefeld University
CeBiTec / Faculty of Technology - Computational Metagenomics
Telephone: +49 521 106-2910
Email: asczyrba@cebitec.uni-bielefeld.de
Dr Andreas Schlüter, Bielefeld University
CeBiTec - Genome Research of Industrial Microorganisms
Telephone: +49 521 106-8757
Email: aschluet@cebitec.uni-bielefeld.de